| |
Spectral Assignment - Applications
& Techniques in NMR Research
Scientists at the NCI-FCRDC and Accelrys have used FELIX
for spectral assignment of NMR data.
Rapid Assignment of Multidimensional NMR Spectra
R. Andrew Byrd and Kristine E. Miller, Macromolecular
NMR Section, ABL-Basic Research Program, NCI-FCRDC,
Frederick, MD.
Reducing Errors in NOE Crosspeak Assignment
Accelrys Inc.
Improved Structure Calculations Through Iterative
Assignment Filtering Using NOESY Back-Calculation
Accelrys Inc.
-----------------------------------------------------------------------------------------------------------------------------------------
Rapid Assignment of Multidimensional NMR Spectra
 23kb
23kb |
Each strip plot is
centered on the amide proton resonance in
the plane of the amide nitrogen and contains
the NOE crosspeaks between the aliphatic and
amide protons used to determine sequential
and long-range assignments among the first
six residues of ubiquitin. |
With the development of heteronuclear double and triple
resonance 3D/4D NMR spectroscopy, the size limit of the
protein, whose structure can be successfully tackled by
NMR, has been continuously increased for the last five
years. In the assignment process of these types of
spectra, automated algorithms and good interactive tools
are crucial. The capabilities within FELIX-Assign for
working with multiple data sets (via separate or
overlaid displays) facilitate the analysis procedure.
Capabilities for shuffling between planes of 3D or 4D
data (indicating whether a crosspeak is at maximum
intensity in the currently displayed or adjacent plane)
and drawing 1D vectors (parallel to any dimension) in
real time vastly improve the efficiency of the analysis.
These features enable the user to conduct the analysis
directly on the workstation screen, without the
time-consuming and cumbersome process of plotting on
paper the many planes within a multidimensional
spectrum.
The interactive ability to dea with both vector- and
contour-mode data is extremely useful for analyzing less
well-behaved data. The automatic tools for matching spin
system patterns to amino acid types and for performing
automatic neighbor finding among the identified spin
systems speeds up the process for completing the
assignments.
FELIX- Assign simplifies the interaction with the
spectra and the resonance and peak databases, which
permits the scientist to focus more on the analysis of
the data (Figure 5 -25kb).
Contributed by Drs. R. Andrew Byrd and Kristine E.
Miller, Macromolecular NMR Section, ABL-Basic Research
Program, NCI-FCRDC, Frederick, MD.
-----------------------------------------------------------------------------------------------------------------------------------------
Reducing Errors in NOE Crosspeak Assignment
 67kb
67kb |
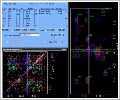
The image depicts a simple-to-use,
menu-driven interface for suggesting and
assigning individual spin frequencies
(resonances) to each dimension of the NOE
crosspeak (upper left). The full NOESY spectrum is
depicted on the lower left with each NOE crosspeak
colored accroding to its current assignment state. The
yellow, purple and cyan lines in the right-hand spectrum
represent the four possible frequencies (two in each
dimension), color-coded based on the spin system to
which they belong. |
The accurate assignment of NOE crosspeaks is critical
for the generation of accurate structures. A single,
erroneous distance restraint can lead to an incorrect
model. FELIX-Assign provides many tools for aiding the
accurate assignment of the NOE crosspeaks (Figure 6
-67kb). For example, the LYS_6:HN assignment can be
built by clicking the mouse on the various lists,
preventing typographical errors and the introduction of
erroneous restraints into the structure calculations.
Additionally, the assignment state of each crosspeak in
the database is stored for later use, such as in
crosspeak coloring for visualizing the progress of the
assignment procedure.
-----------------------------------------------------------------------------------------------------------------------------------------
Improved Structure Calculations Through Iterative
Assignment Filtering Using NOESY Back-Calculation
 63kb
63kb
 107kb
107kb
The quality of NMR-generated structures can be
dramatically improved by using starting models as guides
to identify misassignments and to assign additional
crosspeaks by imposing distance or NOE back-calculated
intensity cutoffs to resolve assignment ambiguities. An
iterative repetition of this cycle leads to more
accurate and precise models. By combining the spectral
visualization capabilities within FELIX-Model and the
spectral simulation methods within NMR Refine, the NOESY
spectrum can be generated from an ensemble of structures
and used to verify and suggest additional assignments.
The left-hand panel of Figure 7 (63kb) depicts the
experimental 2D-NOESY spectra for Human Anaphylatoxin
C3a with the crosspeaks colored according to their
assignment state. The green colored peaks indicate those
crosspeaks that were used as input constraints to DG-II
calculations, and the red colored peaks represent those
with ambiguous assignments. The right-hand panel depicts
the back-calculated spectra simulated from an ensemble
of 10 C3a structures using IRMA. The majority of
individual proton resonances have been previously
assigned(5), and all of the crosspeaks in the
back-calculated spectra have assignments. It is evident
that many more peaks are uniquely assigned in the
back-calculated spectrum. These simulated crosspeak
assignments can be used to verify the uniquely assigned
experimental crosspeaks. They also suggest plausible
assignments for the experimental crosspeaks that have
ambiguous assignments due to resonance and crosspeak
overlap(6).
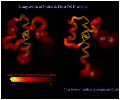
Figure 8 (107kb) depicts the C3a DG-II-generated
structures rendered as variable-width ribbons. The
diameter of the ribbon is proportional to the average
per-residue heavy-atom RMSD over the ensemble. The
backbone atoms of the ensemble were superimposed prior
to the RMS calculation. The two ribbons shown are before
and after several iterations of assignment filtering
using back-calculated 2D-NOESY spectra(7). This analysis
led to an increase in the total number of constraints
(~10%). The largest improvements in the quality of the
structure arose from the removal or recategorization of
erroneous crosspeak assignments. In addition, frequently
a crosspeak in the experimental spectrum that was
assigned as arising from a single proton pair turned out
to contain more than one proton pair within the
simulated spectrum, and thus was removed from the
restraint list.
-----------------------------------------------------------------------------------------------------------------------------------------
Back to top
More Case Studies
|
|
•
Product Overview
•
Felix 2D/ND
•
Modules
 Felix 2D/ND
Felix 2D/ND
 Felix Assign
Felix Assign
 Felix
Autoscreen
Felix
Autoscreen
 Felix Model
Felix Model
•
Case Studies
 Spectral
Assignment
Spectral
Assignment
 Spectral
Processing & Analysis
Spectral
Processing & Analysis
 Structure
Calculation & Analysis
Structure
Calculation & Analysis
•
System Requirements
•
How to Try/Buy Felix |